
Introduction
All current surface exposure/accessibility prediction programs are based on either Emini or Hoop-Woods algorithms. In our experience, both algorithms often miss, or wrongly identify, the exposure of the peptides on the surface of the molecule. Therefore, we have developed EpiQuest A (for accessibility) to be used together with our B-epitope prediction program EpiQuest-B.
As our primary aim is to predict epitope accessibility for an antibody, one way to evaluate the program is to take molecules with known immunodominant epitopes, recognized by antibodies reactive with the intact molecule, and to see, whether the sequence of the epitope will be included with the respective software into surface-exposed or a non-accessible (from the surface) domain.
EpiQuest-B | EpiQuest-A | EpiQuest-IM | EpiQuest-C |EpiQuest-T | EpiQuest-H | EpiQuest-M | In Charge | EpiStat
The algorithm
The algorithm is based on the original values for individual amino acids developed by Janin* transformed into a matrix of probabilities for amino acid exposure at the surface of the molecule. However, in contrast to Emini** algorithm, the actual context (next neighbor) was contributing to the individual values for amino acid probability of being included into the exposed fragment. The algorithm was then trained on actual cases of surface-accessible immunodominant epitopes in protein molecules resulting in the final matrix engaged by the program.
As we are primarily interested in epitope exposure, and their potential for eliciting immune response recognizing the intact molecule, our algorithm is focused on detecting areas that may contain diagnostic epitopes or sequences for preparing antibodies that will be reactive with the native protein molecule.
*Janin, J., S. Wodak, M. Levitt, and B. Maigret. 1978. Conformation of amino acid side-chains in proteins. J. Mol. Biol. 125:357-386.
** J of Virology 1985, p. 836-839
The program output
The program produces two types of results: 1) a histogram of probabilities for particular domains to be accessible at the surface of the native protein, as well as bars indicating continuous domains exposed at the surface that are longer than the predefined size of the peptide (default 8aa's and longer); 2) table of the sequences that are exposed at the surface with the relative values (for the whole sequence and per amino acid) of their probability of exposure.

In the image above, the accessibility profile generated by EpiQuest-N for Ep-CAM molecule is shown. Below are the tabular results, sorted according to the level of SEPR index (the higher the index, the higher the probability for full accessibility of the domain (and the potential epitopes) at the surface.

We would recommend selecting the epitopes that fall within the higher peaks, even if they are relatively narrow. Avoid the long fragments comprised with many short peaks, interrupted (as those in the fragment 68-107): these are usually the highly structured regions with only small fragments of the sequence being properly exposed and accessible for the antibody.
Accessibility and immunogenicity
For immune response against the linear epitope at the molecule's surface (native proteins or their complexes), the accessibility of the sequence is required, but not sufficient. The antigenicity of the sequence is more important, and the overall should be viewed as independent from accessibility. It is the combination of the two that leads to an immunogenic peptide sequence (when the whole molecule is used for vaccination).
We should mention that reviewing accessibility is important when one characterizes the epitopes recognized by antibodies against a pathogen's protein. When testing an immune serum against a collection of overlapping peptides from the sequence of a protein antigen the results may be confusing. Some of the non-accessible peptides may show good recognition by the tested serum. Immune sera often contain IgGs of the non-specified repertoire, and those being quite "sticky"to protein sequences. They may sometimes produce the reaction that may be comparable to a true specific response against the antigen. Therefore, it is important to analyse whether the recognized epitopes are truly accessible at the surface of the native molecule (i.e. diagnostic antigen), or whether they are the hidden ones.
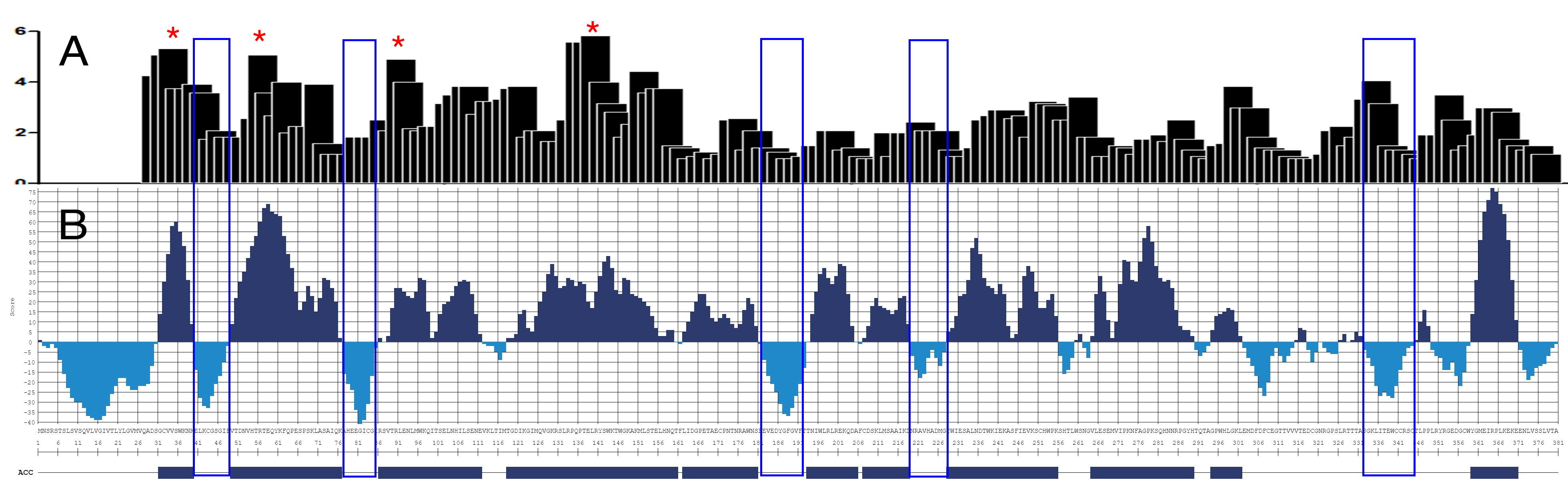
Here we present the reactivity of human sera from dengue-infected patients with the series of overlapping peptides representing the sequence of NS1 protein from Dengue virus 2 (A) and the accessibility profile for this molecule (B). As you may notice, all strong epitopes are within the well-exposed regions of the molecule (marked by *), whereas hidden sequences (framed in blue) do not elicit immune response. However, we can see some response against one of those sequences (the very right frames area). It is advisable to avoid using such sequences as a specific antigen determinant for an antibody assay.
Analysing the Demo sequences
Please see the detailed Manual for EpiQuest-A and Analysis of Demo sequences to master the use of the application.
EpiQuest Suite and site www.epiquest.co.uk belongs to Aptum Biologics Ltd.
EpiQuest® is a registered Trademark of Aptum Biologics Ltd.
© 2018, 2020, 2021 Aptum Biologics Ltd.



