

EpiQuest - InCharge
Determine the pI point of a protein (peptide)
and build the titration curve.


InCharge,isoelectric point,isoelectric point of amino acids,calculating isoelectric point,pi amino acid,pi of proteins,isoelectric point of protein,isoelectric point of lysine,isoelectric point of histidine,isoelectric point of arginine,calculating pi of peptide,calculating pi of amino acid,pi point,glycine isoelectric point,isoelectric point of cysteine,isoelectric point of peptide,isoelectric ph,isoelectric point of aspartic acid,isoelectric point of glutamic acid,pi peptide,glutamic acid isoelectric point,dna isoelectric point,albumin isoelectric point,igg isoelectric point,isoelectric point antibody,isoelectric ph of amino acid,isoelectric ph of protein,isoelectric point of amino acids pdf,isoelectric point of protein pdf,the isoelectric point is,the isoelectric point of a protein is defined as,isoelectric point glycine,calculating isoelectric point of a peptide,isoelectric point of tripeptide,protein a isoelectric point,isoelectric point pdf,carbonic anhydrase isoelectric point,human igg isoelectric point
About InCharge
Prediction of the charge of the molecule, as well as of its isoelectric point (pH at which the charge is neutral) for peptides, small proteins etc. is required for many applications, including peptide/polypeptide conjugation, packaging, delivery, as well as prediction of its mobility in elecrophoresis in 2D gels. The advantage of InCharge is that it is more precise than leading similar software.
EpiQuest-B | EpiQuest-A | EpiQuest-IM | EpiQuest-C |EpiQuest-T | EpiQuest-H | EpiQuest-M | In Charge | EpiStat
Matrix & Algorithm
The program employs a quite standard algorithm for this type of programs, the key difference with analogues is in particular "weight"of specific amino acids in the overall charge of the molecule.
Program output
The output provides the calibration curve for the protein charge at different pH, providing the pI pont and the numerical values for the charge at a particular pH point.

Accuracy of the in silico prediction

Here you see the results for the large selection of proteins showing true and predicted pI points for each of them (the numbers indicate the Accession number).
The results are mainly correct for the proteins in the wide range of the molecular weights.
Below (A) are shown experimental data for a large number of proteins and their pI points as predicted by InCharge and PepCalc, the other similar popular program. The actual points for every protein and the correlation curve are shown, demonstrating that on the overall data the prediction of InCharge is closer to the actual charge of the molecule.
In B, the same correlation curves are shown for all 6 programs tested with this set of proteins (of human and E.coli origin). The colour of the line corresponds to the colour marking each individual program in the table below.

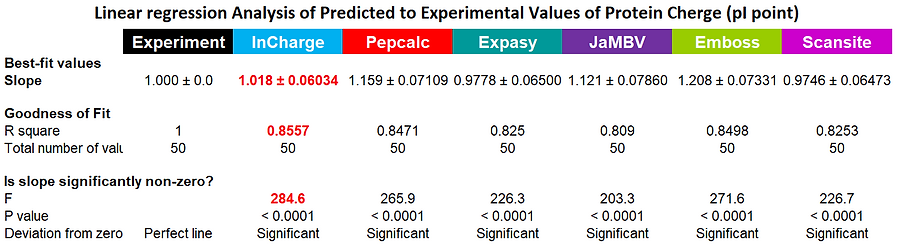
To compare several programs, producing a titration curve and prediction of the pI point for peptides and proteins, we have used 50 proteins (human, mouse, E. coli, S. aureus) with experimentally established pI values. On the right you can see the graphical presentation of the results: individual 50 points were used to create the linear regression line for predicted pI values obtained with each program. Obviously, for experimental data, the predicted and observed results are identical (R square =1); the less accurate the prediction, the smaller the R square will be. As can be seen from both the graphical data, and statistical interpretation of the results, InCharge is substantially more accurate than even closest competitors.

Analysing Demo sequences
You may test the program using our server with a set of demo sequences. For each sequence, the actual reported pI value is shown.
For information about operating the program, please refer to the InCharge Manual, which is also accessible from the program page on the server.
Please note that the program is freely accessible and you may use our server after registering for Demo version of the EpiQuest Suite.
Publications where InCharge was used
Juast a few examples of the published use of InChage in various studies:
Christensen, L.F.B., Malmos, K.G., Christiansen, G., and Otzen, D.E. (2016). A Complex Dance: The Importance of Glycosaminoglycans and Zinc in the Aggregation of Human Prolactin. Biochemistry 55, 3674–3684.
Pavlov, G., and Hsu, J.T. (2018). The pH, temperature, and protein structure effect on β-lactoglobulin A and B separation in anion-exchange chromatography. AIChE Journal 64, 1928–1937.
Serrano, M.A.C., Zhao, B., He, H., Thayumanavan, S., and Vachet, R.W. (2018). Molecular Features Influencing the Release of Peptides from Amphiphilic Polymeric Reverse Micelles. Langmuir 34, 4595–4602.
da Silva, N.R., Ferreira, L.A., Madeira, P.P., Teixeira, J.A., Uversky, V.N., and Zaslavsky, B.Y. (2015). Effect of sodium chloride on solute–solvent interactions in aqueous polyethylene glycol–sodium sulfate two-phase systems. Journal of Chromatography A 1425, 51–61.
EpiQuest Suite and site www.epiquest.co.uk belongs to Aptum Biologics Ltd.
EpiQuest® is a registered Trademark of Aptum Biologics Ltd.
© 2018, 2020, 2021 Aptum Biologics Ltd.
